LDsplit
Introduction
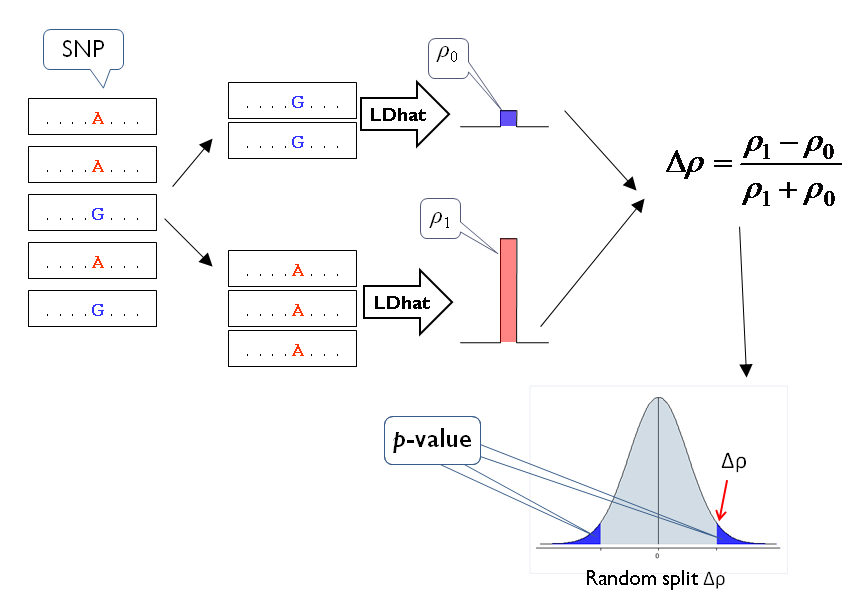
LDsplit is an open source Java program that detects SNPs (single nulceotide polymorphisms) associated with meiotic recombination hotspots. The association is measured by the difference of historical recombination rates at a hotspot between two subpopulations with different alleles at a candidate SNP (Figure 1). With interactive data visualization for exploratory analysis (Figure 2), LDsplit can be used to identify cis-regulatory elements of recombination hotspots both in large scale and high resolution.
Download
LDsplit package in zip file (including executable jar file and Java source code)
User manual of LDsplit (PDF)
Note:
1. After unzipping the LDsplit package (using say WinZip), please do NOT move LDsplit.jar file outside the original folder, because it need to find the lookup files in the "Lookup" folder. For convenient access under Windows, you may create a shortcut to the LDsplit.jar file and paste the shortcut link in any folder you like (such as the desktop).
2. LDsplit takes some time to finish the running of "Calculate recombination profiles". After you load input files, set the parameters and push the "Start" button, the progress bar may appear "frozen" for the first few minutes (depending on input size and parameters), but actually it is running. After a while, the progress bar will show percentage of work already done. Your patience is very much appreciated.
3. LDsplit has been tested under Windows XP/NT/7, and Linux (Unbuntu).

Figure 1. The workflow of LDsplit Algorithm
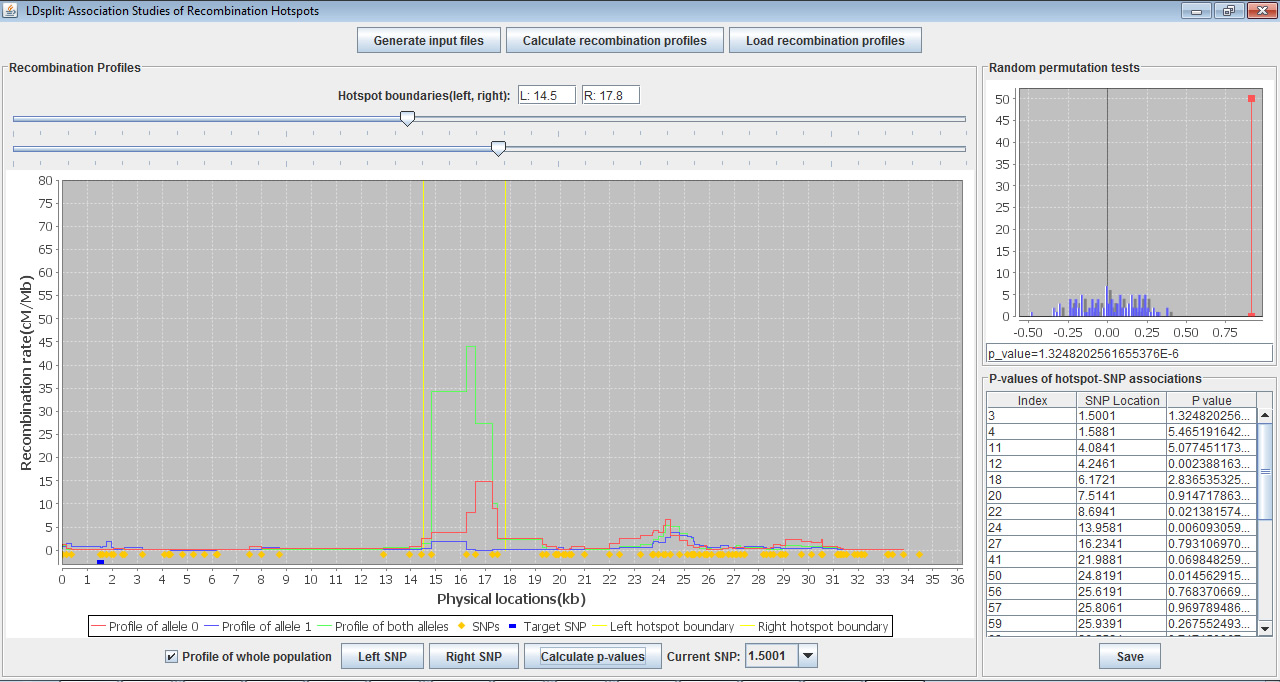
Figure 2. Screenshot of LDsplit interface
Acknowledgements
This project is currently supported by:
Previously it was partially supported by:
References
J. Zheng, PP Khil, RD Camerini-Otero, TM Przytycka. Detecting sequence polymorphisms associated with meiotic recombination hotspots in the human genome. Genome Biology, 11:R103. 2010 (IF = 9.036).
P. Yang, M. Wu, C-K Kwoh, PP Khil, RD Camerini-Otero, TM Przytycka, J. Zheng*. Predicting DNA Sequence Motifs of Recombination Hotspots by Integrative Visualization and Analysis. Proc. International Symposium on Integrative Bioinformatics, pp. 52-58, 2012.
M. Wu, C-K Kwoh, TM Przytycka, J. Li, J. Zheng*. Epigenetic Functions Enriched in Transcription Factors Binding to Mouse Recombination Hotspots. Proteome Science, 10(Suppl 1):S11, 2012 (presented at BIBM'11) (IF = 2.33).
M. Wu, C-K Kwoh, X-L Li, J. Zheng*. NetPipe: A Network-based Pipeline for Discovery of Genes and Protein Complexes Regulating Meiotic Recombination Hotspots. Proc. ACM Conference on Bioinformatics, Computational Biology and Biomedicine (ACM-BCB), Orlando, FL, USA, pp. 20-27, 2012.
M. Wu, C-K Kwoh, TM Przytycka, J. Li, J. Zheng*. Integration of Genomic and Epigenomic Features to Predict Meiotic Recombination Hotspots in Human and Mouse. Proc. ACM Conference on Bioinformatics, Computational Biology and Biomedicine (ACM-BCB), Orlando, FL, USA, pp. 297-304, 2012.
Last update: 11 Nov. 2012